| CATP |
2 x 10-3 M-1 |
| CADP |
2 x 10-4 M-1 |
| CPi |
10-2 M-1
|
| CH+ |
5 x 10-8 M-1(pH
approx. 7.3)
|
The Gibbs energy change under such conditions (temperature 310
oK,
or 37
oC) will be
 =
=  o'
+ 2.3 RT log ( CADP
CPi CH+ / CATP )
= -30 - 19.6 = - 49.6 kJ mol-1
o'
+ 2.3 RT log ( CADP
CPi CH+ / CATP )
= -30 - 19.6 = - 49.6 kJ mol-1
This figure, calculated from the actual concentrations of the
reaction components, reflects the energy available as a driving force
for any other process coupled to ATP hydrolysis under given conditions.
It follows that the same 49.6 kJ mol
-1 must be provided by
the proton transport across the membrane down the electrochemical
gradient to maintain such a high ATP/ADP ratio. If we assume that 3
protons are transported per each ATP molecule synthesized, a
transmembrane H
+ electrochemical gradient of 49.6 / 3
= 16.5
kJ mol
-1
(i.e.,
protonmotive
force of 171 mV) is necessary.

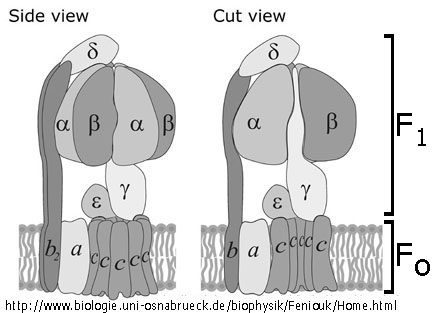
 ).
In case of hydrolysis the enzyme functions as an ATP-driven proton pump
and generates
).
In case of hydrolysis the enzyme functions as an ATP-driven proton pump
and generates  o
o


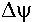 + 2.3
RT (pHP - pHN),
+ 2.3
RT (pHP - pHN), is the
transmembrane electrical potential difference in
volts. The value of
is the
transmembrane electrical potential difference in
volts. The value of 
